Protein methylation is among the most common and important post-translational modifications and plays a major role in several biological processes, including signal transduction, DNA repairing, transcriptional regulation, gene activation, gene repression and RNA processing1. It involves the covalent attachment of a methyl group onto the C-terminus, N-terminus or one of eight amino acid residues. These include arginine, lysine, aspartate, asparagine, glutamine, glutamate and histidine1,2. The transfer of one to three methyl groups from S-adenosyl-methionine onto the substrate protein is catalysed by diverse methyltransferases1–5. Methyltransferases generally use base catalysis in order to create reactive nucleophils which then attack the methyl group of S-adenosyl-methionine in an SN2 nucleophilic substitution reaction [5]. S-adenosyl-L-homocysteine is released in the process. The methyl group(s) can be removed again by demethylases1–4. Due to mentioned amino acid residues, it is self-evident that the methyl moieties are bound to different atoms and therefore are classified into N-methylation, O-methylation and S-methylation, whereby N-methylation is the most abundant1. A residue can be methylated a maximum of three times. Methylation mainly occurs on lysine and arginine residues1–4,6. Other post-translational modifications at the same site can be suppressed by methylation4. This modification has a relatively small influence on the physico-chemical properties of substrate proteins1.
Monomethylation
Overview
Protein methylation, catalyzed by highly specific methyltransferase enzymes, is a relatively common post-translational modification and plays an important role in various biological processes. Monomethylation occurs most frequently at arginine and lysine residues.
| pKa | NC | Loss | Gain | Deltamass | H | AA | UV-Spec | Pattern |
| – | No | H | CH3 | Av: 14.0266 M: 14.0157 | – | C, D, E, H, K, R, S, T, ^, $ | – | – |
In-depth mechanism
Monomethylation can occur on all eight mentioned amino acid residues (arginine, lysine, aspartate, asparagine, glutamine, glutamate and histidine) as well as N-terminus and C-terminus. In monomethylation, a methyl group is transferred from S-adenosyl-methionine to a terminal nitrogen, oxygen or sulfur atom. In the case of lysine, the transfer is performed by protein lysine methyltransferase (PKMT) and in the case of arginine by protein arginine methyl transferase (PRMT) type I and type II1–4,6. The mechanism of monomethylation is shown in figure 1 using the example of lysine and arginine. Monomethylation increases the molecular mass by 14 Da of the modified protein1,2.
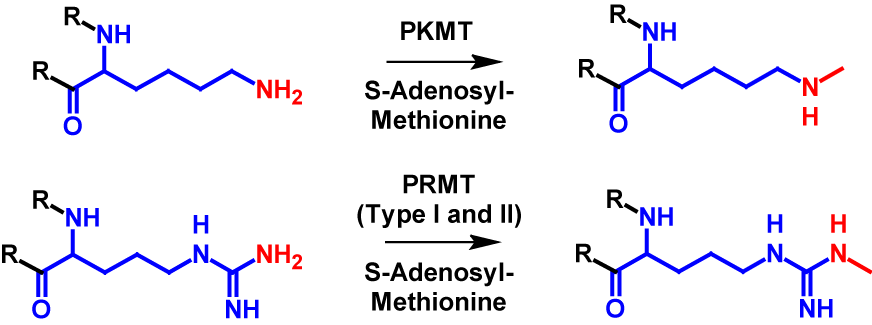
Dimethylation
Overview
Protein methylation, catalyzed by highly specific methyltransferase enzymes, is a relatively common post-translational modification and plays an important role in various biological processes. Dimethylation occurs most frequently at arginine and lysine residues.
| pKa | NC | Loss | Gain | Deltamass | H | AA | UV-Spec | Pattern |
| – | No | H2 | C2H6 | Av: 28.0532 M: 28.0313 | – | K, R, ^ | – | – |
In-depth mechanism
Dimethylation occurs at significantly fewer sites than monomethylation. It mainly occurs at arginine and lysine residues1–4 but can also occur at the N-terminus7. In dimethylation, two methyl groups are transferred from S-adenosyl-methionine onto a nitrogen atom of the receiving residue. For lysine the transfer is catalysed by PKMT, the same enzyme as monomethylation. For dimethylated arginine residues exist two different structures. The guanido group can either be methylated symetrically or asymetrically. The methylation of symmetric dimethylarginine is catalysed by PRMT type I and the methylation of asymmetric dimethylarginine is catalysed byPKMT type II1–4. The mechanism of dimethylation is shown for lysine and arginine in figure 2. Dimethylation increases the molecular mass by 28 Da of the modified protein1,2.
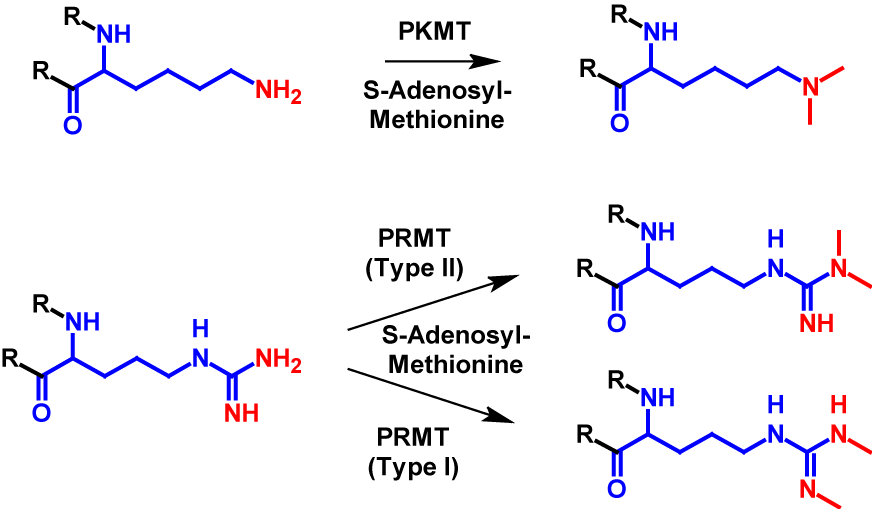
Trimethylation
Overview
Protein methylation, catalyzed by highly specific methyltransferase enzymes, is a relatively common post-translational modification and plays an important role in various biological processes. Trimethylation occurs most frequently at lysine residues.
| pKa | NC | Loss | Gain | Deltamass | H | AA | UV-Spec | Pattern |
| – | No | H3 | C3H9 | Av: 28.0532 M: 28.0313 | – | K, ^ | – | – |
In-depth mechanism
Lysine residues in proteins are the only amino acids that can be trimethylated1–4. Additionally, the N-terminus is a possible site for trimethylation, however lysine trimethylation is more frequent7,8. Trimethylation results in the N-terminus or the lysine residue being permanently positively charged [4]. The mechanism of lysine trimethylation is shown in figure 3. PKMT again catalyses the transfer of a methyl group from S-adenosyl-methionine onto the lysine residue. Trimethylation increases the molecular mass by 42 Da of the modified protein1,2.
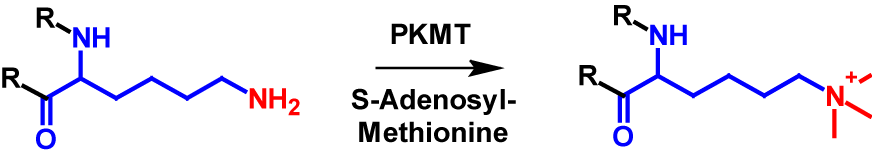
References
- 1.Wang Q, Wang K, Ye M. Strategies for large-scale analysis of non-histone protein methylation by LC-MS/MS. The Analyst. 2017;142:3536–3548. doi:10.1039/c7an00954b
- 2.Afjehi-Sadat L, Garcia BA. Comprehending dynamic protein methylation with mass spectrometry. Current opinion in chemical biology. 2013;17:12–19. doi:10.1016/j.cbpa.2012.12.023
- 3.Wang Y-C, Peterson SE, Loring JF. Protein post-translational modifications and regulation of pluripotency in human stem cells. Cell Research. 2014;24:143–160. doi:10.1038/cr.2013.151
- 4.Erce MA, Pang CNI, Hart-Smith G, Wilkins MR. The methylproteome and the intracellular methylation network. Proteomics. 2012;12:564–586. doi:10.1002/pmic.201100397
- 5.Jarrett JT. Surprise! A hidden B12 cofactor catalyzes a radical methylation. Journal of Biological Chemistry . 2019;294:11726–11727. doi:10.1074/jbc.H119.009976
- 6.Yang Y, Bedford MT. Protein arginine methyltransferases and cancer. Nature Reviews Cancer. 2013;13:37–50. doi:10.1038/nrc3409
- 7.Stock A, Clarke S, Clarke C, Stock J. N-terminal methylation of proteins: structure, function and specificity. FEBS letters. 1987;220:8–14. doi:10.1016/0014-5793(87)80866-9
- 8.Falnes P., Jakobsson ME, Davydova E, Ho A, Maecki J. Protein lysine methylation by seven-β-strand methyltransferases. Biochemical Journal. 2016;473:1995–2009. doi:10.1042/BCJ20160117