Overview
O-Sulfation is a common post-translational modification of tyrosine residues in eukaryotes, but has not been observed in yeast and prokaryotes. Sulfation is limited to secretory and trans-membrane proteins that have passed the trans-Golgi network, where two membrane-bound tyrosylprotein sulfotransferase enzymes catalyze the transfer of sulfate from adenosine 3’-phosphate5’-phosphosulfate to the tyrosine phenol.
| pKa | NC | Loss | Gain | Deltamass | H | AA | UV-Spec | Pattern |
| Acidic -3.88 | No | H | SO3H | Av: 80.063 M: 79.9568 | – | Y | – | – |
In-depth mechanism
O-Sulfation is a post-translational modification in which a sulfuryl group is covalently linked tothe hydroxyl group of a tyrosine residue1–5. The sulfuryl group must be activated first inorder for the transfer to take place. An active sulfuryl group is formed by the reaction of inorganic sulfate with ATP resulting in adenosine-5’-phosphosulfate (APS) and 3’-phospho-adenosine-5’-phosphosulfate (PAPS). These two molecules are known as universal sulfate donors1–4. In fungi and few bacteria PAPS is preferred as donor while in plants, algae and most bacteria APS is preferred [63]. The transfer is catalysed by tyrosylprotein sulfotransferases (TPST) which aremembrane-associated proteins located in the trans-Golgi network1–3,5. The mechanism of O-sulfation is shown in figure 28.
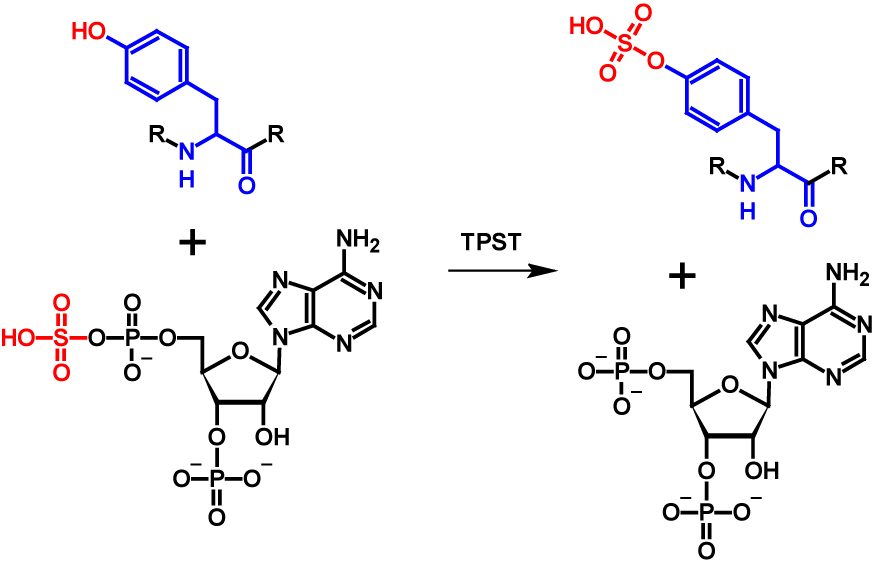
O-Sulfation occurs in higher eukaryotic species. So far it has been observed in all tested mammalian cell lines, but not in prokaryotes or yeast2. Thus far, no specific consensus sequence for O-sulfation sites has been reported. However, sulfated tyrosine residues are surrounded by many acidic amino acids residues2,6. The function of O-sulfation is unknown for most proteins. The function of the majority of known tyrosine-sulfated proteins seems to be the participation in protein-protein interaction, which is at least partially regulated by the sulfuryl group1–3. In general, O-sulfation does not seem to be essential for the proper function. Enzyme activity is reduced in absence, but not entirely eliminated. At the current time, O-sulfation has been observed only in secreted and transmembrane proteins1,2. Tyrosine O-sulfation increases the molecular mass of the modified protein by 80 Da5. In addition, the pKa value is lowered from 10.07 of tyrosine to -3.88 for phenylsulfate. Furthermore, the possible sites for hydrogen bonds are quadrupled in tyrosine-sulfated proteins7.
References
- 1.Moore KL. The Biology and Enzymology of Protein Tyrosine O-Sulfation. Journal of Biological Chemistry . 2003;278:24243–24246. http://www.jbc.org/content/278/27/24243.short
- 2.Walsh G, Jefferis R. Post-translational modifications in the context of therapeutic proteins. Nature Biotechnology. 2006;24:1241–1252. doi:10.1038/nbt1252
- 3.Yang Y-S, Wang C-C, Chen B-H, Hou Y-H, Hung K-S, Mao Y-C. Tyrosine sulfation as a protein post-translational modification. Molecules (Basel, Switzerland). 2015;20:2138–2164. doi:10.3390/molecules20022138
- 4.Woods AS, Wang H-YJ, Jackson SN. Sulfation, the Up-and-Coming Post-Translational Modification: Its Role and Mechanism in Protein−Protein Interaction. Journal of Proteome Research. 2007;6:1176–1182. doi:10.1021/pr060529g
- 5.Zhao J, Saunders J, Schussler SD, et al. Characterization of a novel modification of a CHO-produced mAb: Evidence for the presence of tyrosine sulfation. mAbs. 2017;9:985–995. doi:10.1080/19420862.2017.1332552
- 6.Kanan Y, Al Ubaidi MR. Detection of Tyrosine Sulfation on Proteins. Current Protocols in Protein Science. 2015;80:14.7.1–14.7.20. doi:10.1002/0471140864.ps1407s80
- 7.Balsved D. Hydrolysis of Tyrosine Sulfate in Acidic Solutions and the Occurrence of TyrosineSulfation in the G Protein-Coupled Receptor PAC1. Published online 2006.