Glycosylations should not be neglected for the correct calculation of the molecular mass, the isoelectric point and the mass-specific UV absorption coefficient. Therefore Prot pi provides a tool to draw glycans as a posttranslational modification of proteins. This short guide deals with how to add two complex-type N-Glycosylation G1 with a sialic acid (N-acetylneuraminic acid) to the amino acid sequence of the heavy chains (HC) of a monoclonal antibody. Even though, in most cases, immunoglobulin G (IgG) is additionally modified, no further posttranslational modifications were described in this article.
How-to
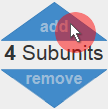
First of all go to the Protein Tool of Prot pi and enter the amino acid sequence of your protein. For this tutorial the sequence of Canakinumab, a recombinant human anti-human-IL-1β IgG, from DrugBank.ca was used. The IgG is composed of four subunits – two identical heavy chains (HC) and two identical light chains (LC). Therefore add three subunits to a total of four subunits by clicking three times the “add” button (figure 1).
Enter the headline (starting with a “>”) into the first and the second text area followed by a “return”. The headlines are optional, but are helpful for the orientation in the results. Then copy the amino acid sequence of the HC on a new line below the headline in the first and the second text area.
Heavy chain:
>8836_H|canakinumab|Homo sapiens||H-GAMMA-1 (VH(1-118)+CH1(119-216)+HINGE-REGION(217-231)+CH2(232-341)+CH3(342-448))|||||||448||||MW 49253.6|MW 49253.6| QVQLVESGGGVVQPGRSLRLSCAASGFTFSVYGMNWVRQAPGKGLEWVAIIWYDGDNQYY ADSVKGRFTISRDNSKNTLYLQMNGLRAEDTAVYYCARDLRTGPFDYWGQGTLVTVSSAS TKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGL YSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKRVEPKSCDKTHTCPPCPAPELLGGPS VFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYNST YRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTLPPSREEMT KNQVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQ GNVFSCSVMHEALHNHYTQKSLSLSPGK
Light chain:
>8836_L|canakinumab|Homo sapiens||L-KAPPA (V-KAPPA(1-107)+C-KAPPA(108-214))|||||||214||||MW 23357.9|MW 23357.9| QVQLVESGGGVVQPGRSLRLSCAASGFTFSVYGMNWVRQAPGKGLEWVAIIWYDGDNQYY ADSVKGRFTISRDNSKNTLYLQMNGLRAEDTAVYYCARDLRTGPFDYWGQGTLVTVSSAS TKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGL EIVLTQSPDFQSVTPKEKVTITCRASQSIGSSLHWYQQKPDQSPKLLIKYASQSFSGVPS RFSGSGSGTDFTLTINSLEAEDAAAYYCHQSSSLPFTFGPGTKVDIKRTVAAPSVFIFPP SDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQDSKDSTYSLSSTLT LSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
Copy the headline and the amino acid sequence of the LC in the same matter into the third and the fourth text area (figure 2).
Now the glycosylation is being defined. Choose “N-Glycosylation” within the “Modifications” tab and press the button “Add Modification” (figure 3).

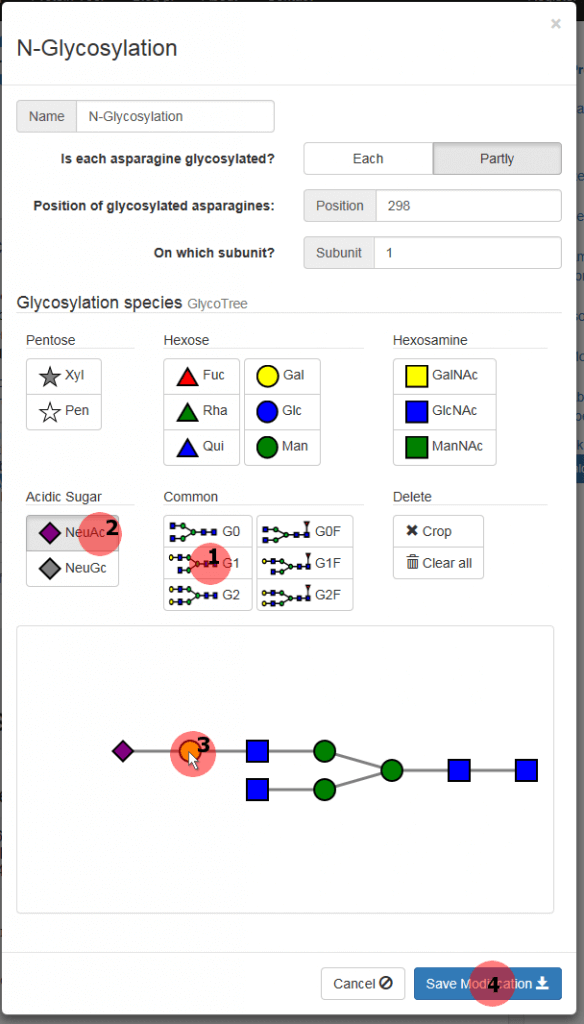
A new window is popped up where you can specify the glycosylation (figure 4). As only the asparagine at the position 298 on the HC is glycosylated, select “Partly” option. Now you can enter the position (298) and the subunit (1) into the appropriate box.
Note that at this point only the glycosylation of one subunit can be defined. An additional modification must be added later for the glycosylation of the second subunit.
Click the “G1” button (1) in the category “Common” and a complex type G1 glycosylation appears in the field below. Then select the “NeuAc” button (2) in the “Acidic Sugar” category. Now an N-acetylneuraminic acid will be appended to the monosaccharide that you click. So click the leftmost galactose (yellow circle) of the glycan tree (3) to get a G1NeuAc glycan. Apply this glycosylation to the N(298) of the HC1 by pressing the “Save Modification” button (4).
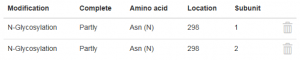
Add now the glycosylation of the second HC in exactly the same manner but for subunit 2. After saving the second glycosylation, the table in the tab “Modifications” should look this way:
With pressing the “Calculate” button you can start the calculation of the physico-chemical parameters of the glycosylated IgG. All physico-chemical parameters are now calculated taking account of the defined N-glycosylation.
Note: Cysteine side chain sulfhydryl groups mostly form disulfide bonds in monoclonal antibodies. And in IgG, normally, the C-terminal lysine of the HC is cleaved during production and N-terminal glutamine appears as pyroglutamic acid. Therefore these modifications must be defined additionally for a correct calculation. Although this is not described in this article, further modifications can be applied in the same manner as the N-glycosylation.
[ratings]
Hi Roland, very nice tool!
Is there a possibility to calculate automatically different variants of modifications? For example 0 to 2 K clipped off? or 0 to 4 NeuAc?
thx for your help! patrizia
Hi Patrizia,
thank you for your comment. Unfortunately there is no possibility to define a range of modifications. You have actually to calculate each of the variants separately. What would be the benefit of the calculation of a variable number of modifications? This would give you a set of results for each variant. Do you need a graphical overlay of the results, or would it be only to save time for entering data?
Cheers,
Roland
Thx for your fast reply!
It would mainly save time! but it would also be interesting to see the influence of the variant as overlay of titration curves. Is it possible to export the titration curve (csv or similar) to do it by myself?
Patrizia
A tool to create an exportable and printable report is under construction. Up until then, you can extract the data for all graphs out of the site source code. The values are rendered in the DOM and if you are familiar with JavaScript, you can easily copy them. Please just let me know, if you need assistance with this.
Cheers,
Roland
It took a while, but now you can download the raw data of all graphics as CSV and image files.
Perfect, thanks a lot!
Patrizia
Dear Roland
I’m working on maillard reaction products. Is there any way to calculate the effect of glycosylation on lysin residue on isoelectric point?
Dear Mahdi Yes, that’s possible. You can define a user-defined modification by selecting “Custom” within the modifications tab. There you can specify if all lysines are glycosylated or (more likely) select “Partly” and enter the position of the modified lysine. Leave the pKa values of modification empty, as Long as you have no sialic acid in your glycosylation. Important: Select the “Hide” option to remove the native charge of lysine since the amine group loose its charge during the maillard reaction. To calculate the correct molecular mass, enter H2O in “Lost elements” and the molecular formula of the glycan (e.g… Read more »
Dear Mahdi
It took a while, but now we have redesigned our algorithm for the modifications. Glycation of a protein with glucose is now a predefined modification. By the way, it doesn’t matter if the protein is glycated by glucose or fructose.
Roland