Overview
S-Nitrosylation is a post-translational modification in which a nitric oxide molecule is bound via a reactive thiol group of a cysteine residue. S-nitrosylation has various regulatory roles in bacteria, yeasts, plants and mammalian cells.
| pKa | NC | Loss | Gain | Deltamass | H | AA | UV-Spec | Pattern |
| – | No | H | NO | Av: 28.9982 M: 28.9902 | – | C | Yes | – |
In-depth mechanism
S-Nitrosylation is a post-translational modification in which a nictric oxide (NO) group is coupled to a thiol group of a cysteine residue resulting in a S-nitrosothiol group1–5. NO is an important short-lived physiological messenger that is highly reactive1,2. It is generated through the conversion of L-arginine to L-citrulline and catalysed by the enzyme NO synthase2,3. The signaling of NO can be categorised in classical and non-classical ways. In the classical way, NO binds to the heme group of guanylyl cyclase, which stimulates the transformation from GTP to cGMP. cGMP is a second messenger, which in turn activates cGMP-dependent protein kinase. cGMP-dependent protein kinase reduces the concentration of potassium and calcium ions in the cytosol. This causes a hyperpolarization of the membrane potential which triggers neurotransmission and vasodilation. The non-classical way describes the process of NO signaling via covalent post-translational modifications which includes among others S-nitrosylation3. The frequency of S-nitrosylation depends on the local concentration of NO and affects intracellular traffic processes, protein phosphorylation and protein-protein interactions3–5. Enzymes do not catalyse the reaction from the thiol to the S-nitrosothiol. However, the denitrosylation is regulated by two enzymes called S-nitrosoglutathione reductase and thioredoxin. If these two enzymes are lacking or inhibited, high concentrations of S-nitrosylated proteins occur2–4. NO concentrations above 100 nM induce S-nitrosylation4. The mechanism of S-nitrosylation is shown in figure 1.
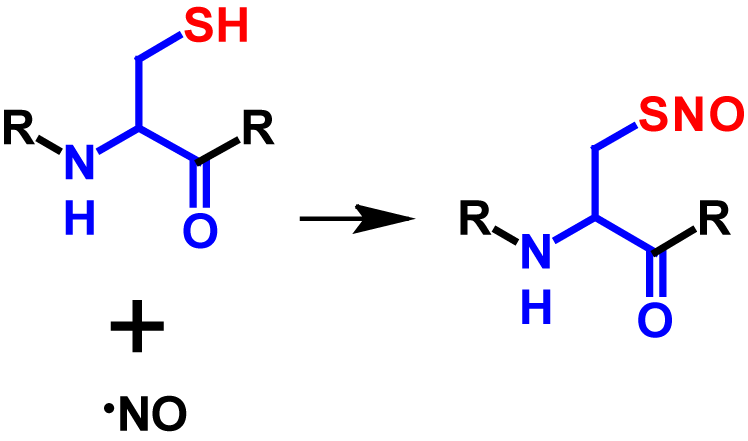
Although S-nitrosylation is a non-enzymatic reaction (except prokaryotes), it is a highly selective modification and is dependent on different factors3,5. Target cysteine residues must be in close proximity of the NO source, within a I/L-X-C-X2-D/E motif as well as within a highly hydrophobic region formed by tertiary protein structure or membranes. In addition, the environment is decisive whether a protein is S-nitrosylated or not. Since a thiolate is required for S-nitrosylation, the reaction is strongly dependent on the neighbouring amino acids, as these strongly influence the pKa of cysteine. Furthermore, S-nitrosylation is hindered by bulky amino acid residues such as phenylalanine, tyrosine, arginine and leucine. Over 3000 proteins with potential S-nitrosylation sites have been identified2,3. S-Nitrosylation results in a molecular mass increase of 29 Da2. Furthermore, the nitroso group causes an additional absorption in the range between 274 – 442 nm6, the maximum being at 336 nm7. The influence of the nitroso group on the absorbance was determined by the difference of the UV spectra of glutathione and S-nitrosoglutathione.
References
- 1.Iwakiri Y, Satoh A, Chatterjee S, et al. Nitric oxide synthase generates nitric oxide locally to regulate compartmentalized protein S-nitrosylation and protein trafficking. Proceedings of the National Academy of Sciences. 2006;103:19777 LP – 19782. doi:10.1073/pnas.0605907103
- 2.Lamotte O, Bertoldo JB, Besson-Bard A, et al. Protein S-nitrosylation: specificity and identification strategies in plants. Frontiers in chemistry. 2014;2:114. doi:10.3389/fchem.2014.00114
- 3.Fernando V, Zheng X, Walia Y, Sharma V, Letson J, Furuta S. S-Nitrosylation: An Emerging Paradigm of Redox Signaling. Antioxidants (Basel, Switzerland). 2019;8. doi:10.3390/antiox8090404
- 4.Ehrenfeld P, Cordova F, Duran WN, Sanchez FA. S-nitrosylation and its role in breast cancer angiogenesis and metastasis. Nitric Oxide. 2019;87:52–59. doi:https://doi.org/10.1016/j.niox.2019.03.002
- 5.Nakamura T, Prikhodko OA, Pirie E, et al. Aberrant protein S-nitrosylation contributes to the pathophysiology of neurodegenerative diseases. Neurobiology of disease. 2015;84:99–108. doi:10.1016/j.nbd.2015.03.017
- 6.Hwang S, Meyerhoff ME. Organoditelluride-mediated catalytic S-nitrosothiol decomposition. Journal of Materials Chemistry. 2007;17:1462–1465. doi:10.1039/B700375G
- 7.Gordon JL, Reynolds MM, Brown MA. Nitric Oxide as a Potential Adjuvant Therapeutic for Neuroblastoma: Effects of NO on Murine N2a Cells. Veterinary Sciences. Published online April 23, 2020:51. doi:10.3390/vetsci7020051